TAGMe DNA Methylation Detection Kits (qPCR) for Urothelial Cancer
PRODUCT FEATURES
Precision

Validated over 3500 clinical samples in double-blind multi-center studies, the product has a specificity of 92.7% and a sensitivity of 82.1%.
Convenient

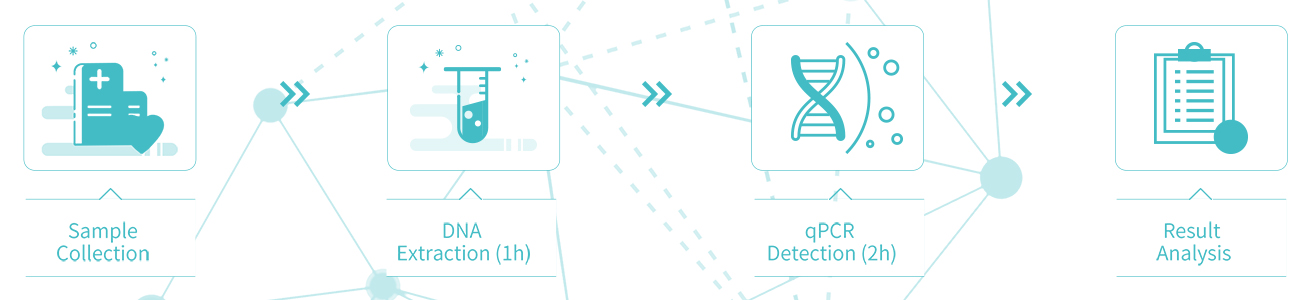
The original Me-qPCR methylation detection technology can be completed in one step within 3 hours without bisulfite transformation.
Non-invasive

Only 30 mL of urine sample is required to detect 3 types of cancer, including renal pelvis cancer, ureteral cancer, bladder cancer at the same time.
Application scenarios
Auxiliary diagnosis: Patients with urothelial cancer can be screened out in a non-invasive manner to assist in clinical diagnosis.
Surgery/chemotherapy efficacy assessment: Evaluate the efficacy of surgery/chemotherapy to assist clinical improvement of therapeutic effect.
Postoperative population recurrence monitoring: Patients with urothelial cancer can be monitored for recurrence in a non-invasive manner non-invasive manner, which improves patient compliance.
Sample collection
Sampling method: Sampling method: Collect a urine sample (morning urine or random urine), add urine preservation solution and mix well, store it at room temperature and label it for the following examination.
Preservation of samples: Samples can be stored at room temperature for up to 14 days, at 2-8 ℃ for up to 2 months, and at -20±5℃ for up to 24 months.
Detection process:3 Hours (Without manual process)
DNA Methylation Detection Kits (qPCR) for Urothelial Cancer

|
Clinical application |
Clinical auxiliary diagnosis of urothelial cnacer; surgery/chemotheray treatment efficacy assessment; postoperative recurrence monitoring |
|
Detection gene |
UC |
|
Sample type |
Urine exfoliated cell sample (urine sediment) |
|
Test method |
Fluorescence quantitative PCR technology |
|
Applicable models |
ABI7500 |
|
Packing specification |
48 tests/kit |
|
Storage Conditions |
Kit A should be stored at 2-30 ℃Kit B should be stored at -20±5℃
Valid for up to 12 months. |